
[57] A Structure-Aware Generative Framework for Exploring Protein Sequence and Function Space
D Shukla, J Martin, F Morcos, DA Potoyan*
Biorxiv (2025)

[56] Backbone Rigidity Encodes Universal Viscoelastic Signatures in Biomolecular Condensates
S Biswas, S Yang, DA Potoyan*
Biorxiv (2025)

[55] Histone H3 tail charge patterns govern nucleosome condensate
formation and dynamics
EF Hammonds, A Singh, KK Suresh, S Yang, SSM Zahorodny, R Gupta,
DA Potoyan* PR Banerjee*, EA Morrison*
Biorxiv (2025)

[54] Molecular Drivers of RNA phase separation
V Ramachandran, DA Potoyan*
Proc Natl Acad Sci (2025)

[53] Chromatin binding regulates phase behavior and morphology
of condensates formed by prion-like
domains
A Suprakar, RB Davis, S Biswas, S Yang,
DA Potoyan* PR Banerjee*
J Mol Biol (2025)
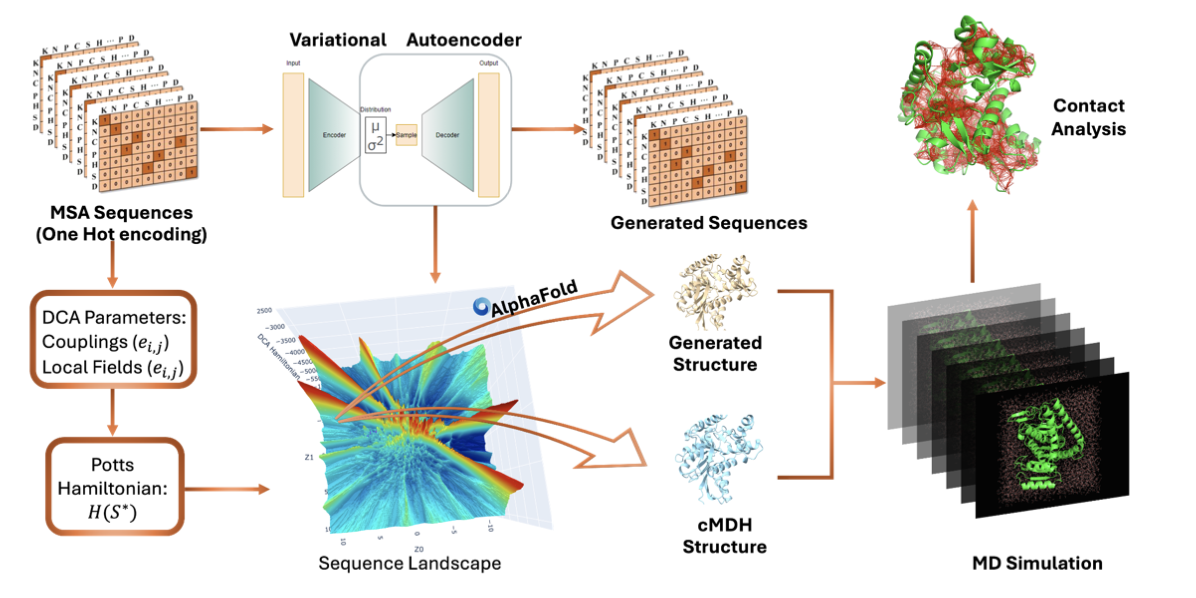
[52] Thermal Adaptation of Cytosolic Malate Dehydrogenase
Revealed by Deep Learning and Coevolutionary Analysis
D Shukla, J Martin, F Morcos, DA Potoyan*
J Chem Theor Comp (2025)
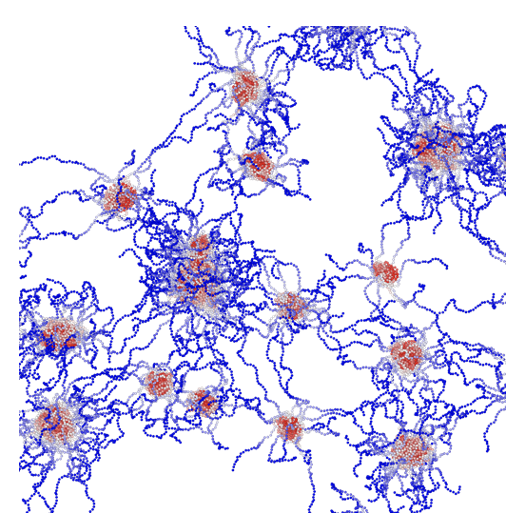
[51] Decoding the dynamics of biomolecular condensates:
An energy landscape approach
S Biswas, DA Potoyan*
PLoS Comp Bio (2025)

[50] Thermal Adaptation of Extremozymes:
Temperature-Sensitive Contact Analysis of Serine Proteases
DP Kulathunga, DA Potoyan*
Biophys J (2025)
(2024)
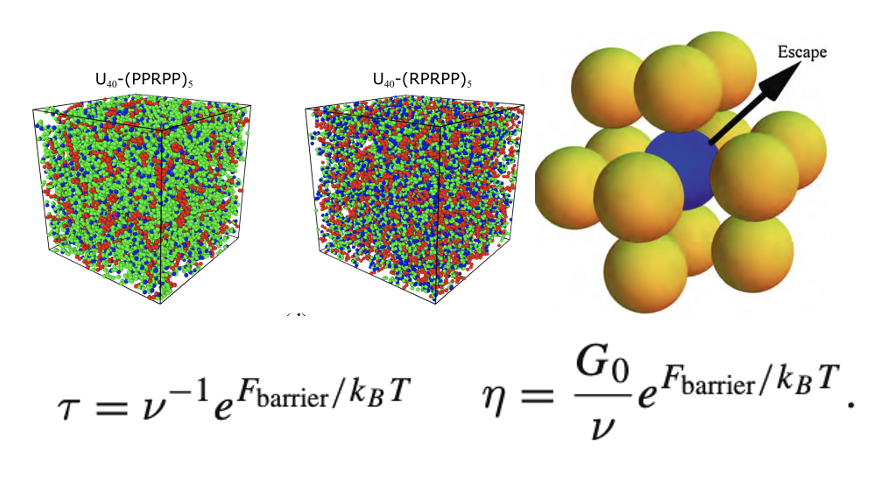
[49] On microscopic origins of flow activation energy in biomolecular condensates
S Yang, DA Potoyan*
J Phys Chem B (2024)
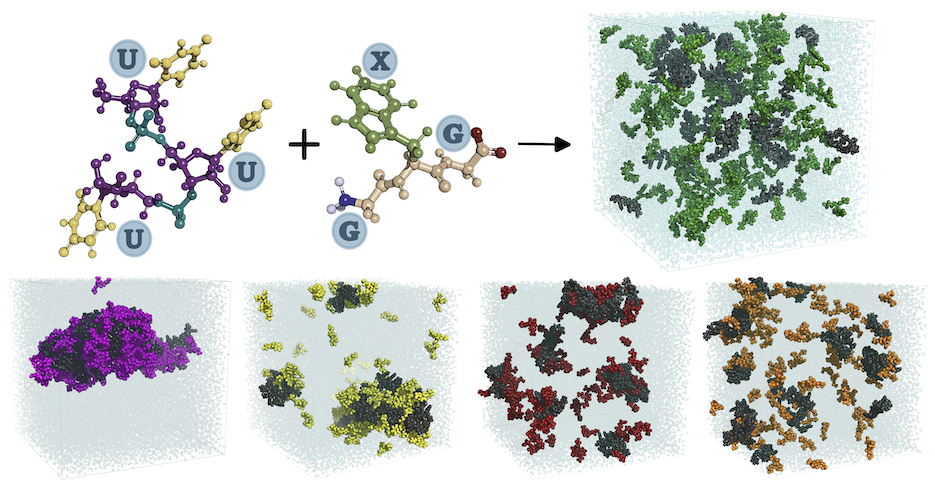
[48] Nucleoprotein phase-separation affinities revealed via
atomistic simulations of short peptide and RNA fragments
V Ramachandran, W Brown, C Gayvert, DA Potoyan*
J Phys Chem Lett (2024)
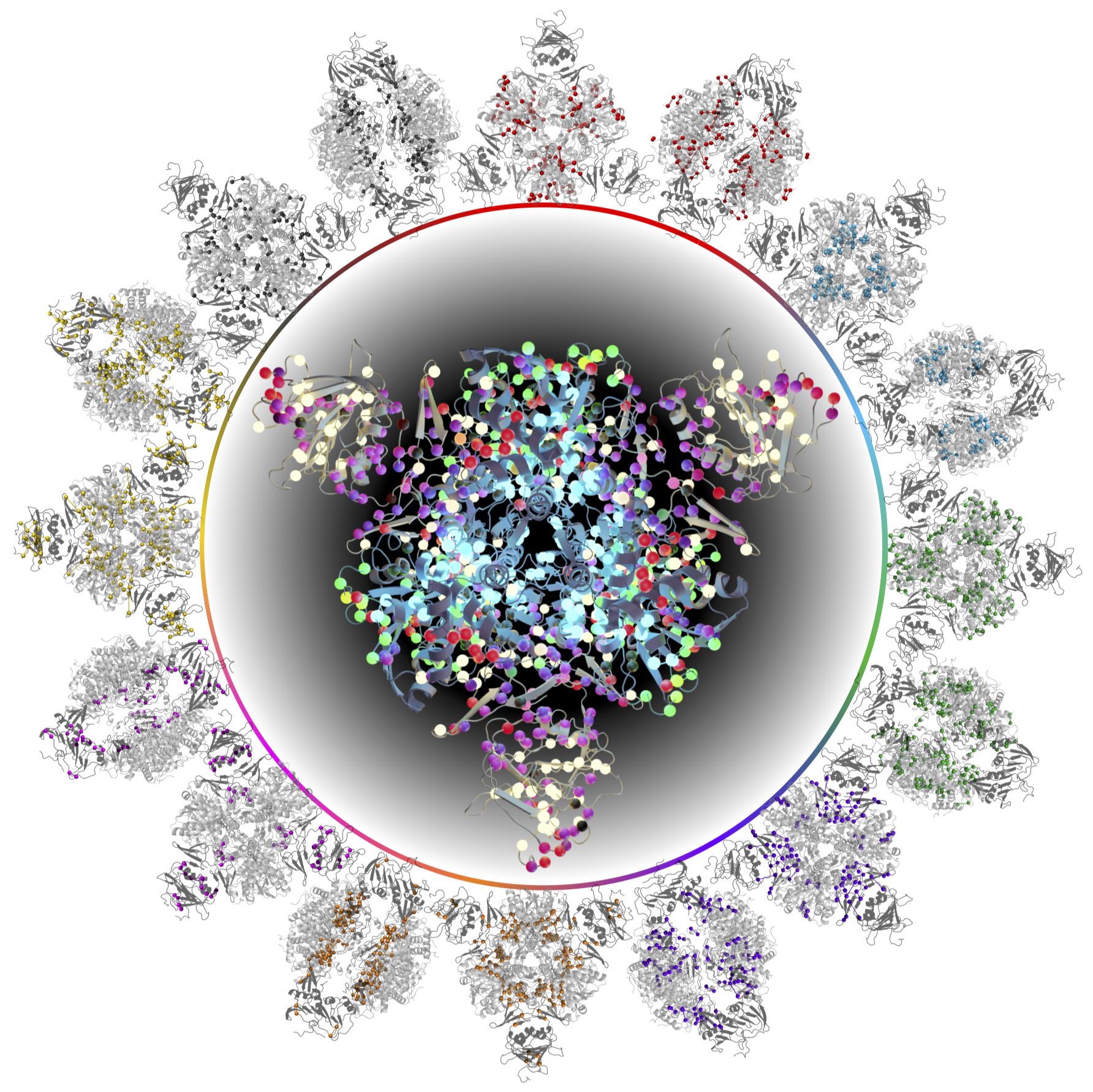
[47] Illuminating protein allostery by Chemically Accurate Contact
Response Analysis
D Burns V Venditti*, DA Potoyan*
J Chem Theor Comp (2024)
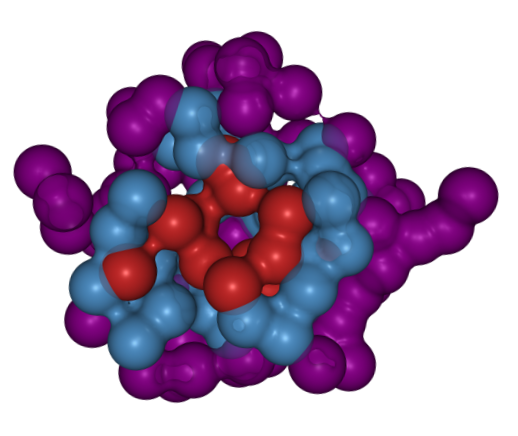
[46] Molecular drivers of aging in biomolecular condensates:
Desolvation, rigidification, and sticker lifetimes
S Biswas DA Potoyan*
Phys Rev X Life (2024)
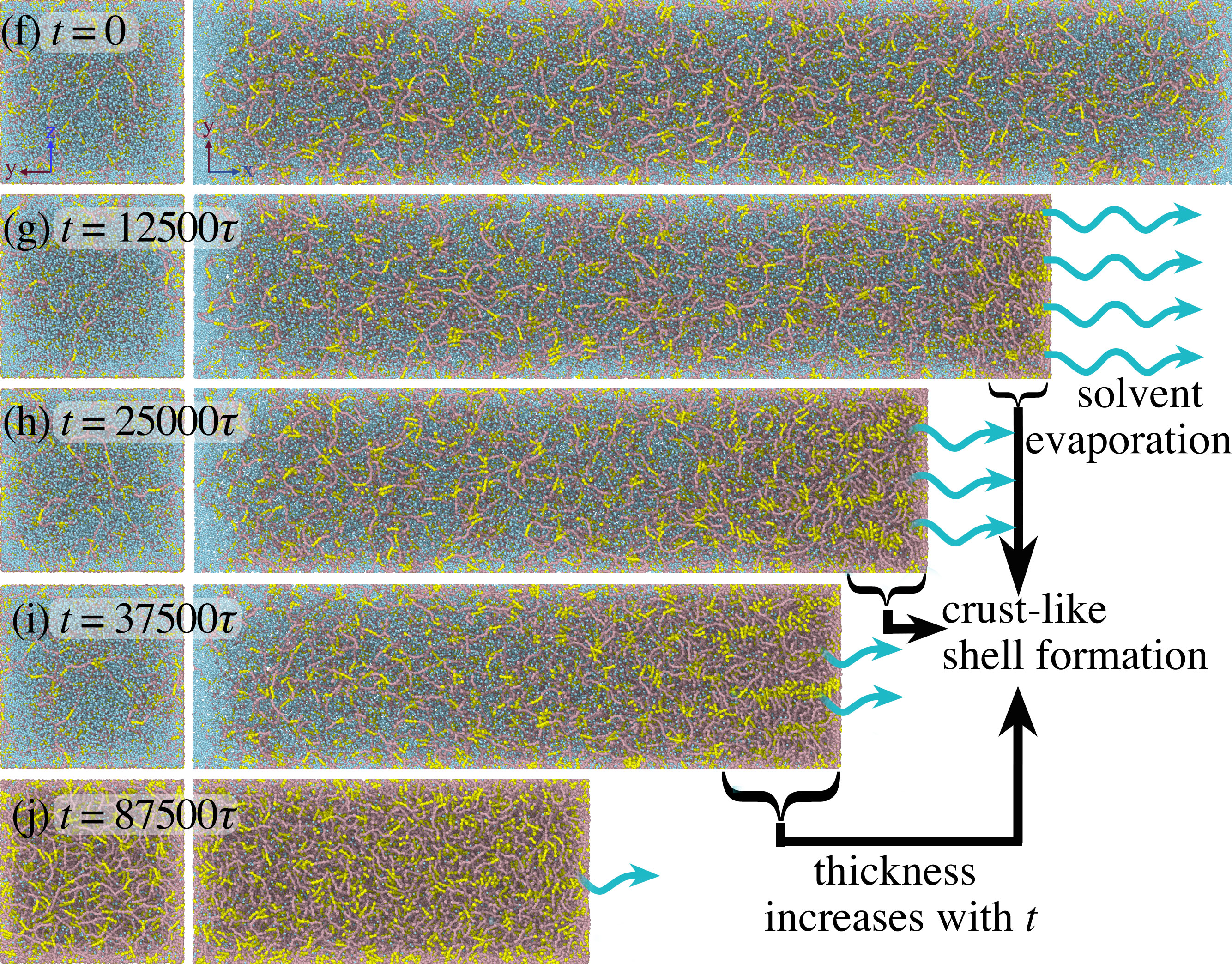
[45] 4D Mesoscale liquid model of nucleus resolves chromatin’s radial organization
R Laghmach, M Di Pierro DA Potoyan*
Phys Rev X Life (2024)
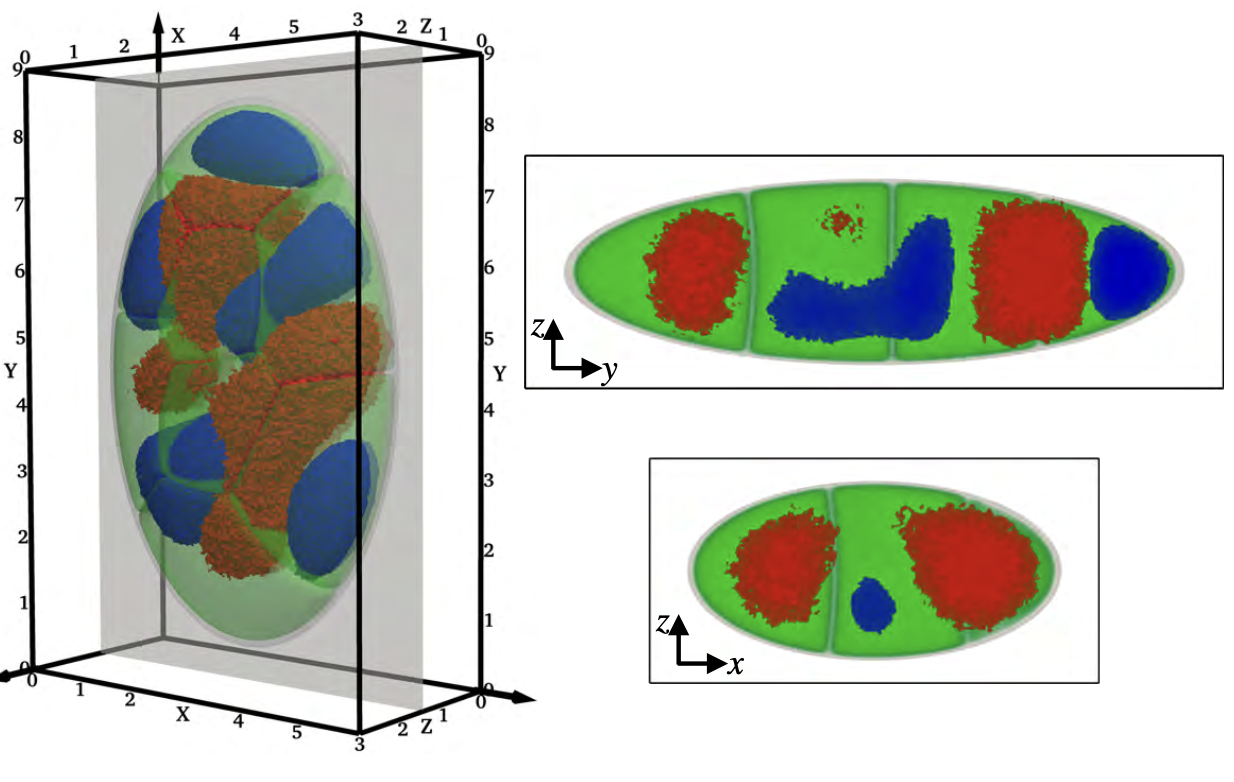
[44] Phase separation of multicomponent peptide mixtures into dehydrated clusters with hydrophilic cores
W Brown, DA Potoyan*
Biophys J (2024)
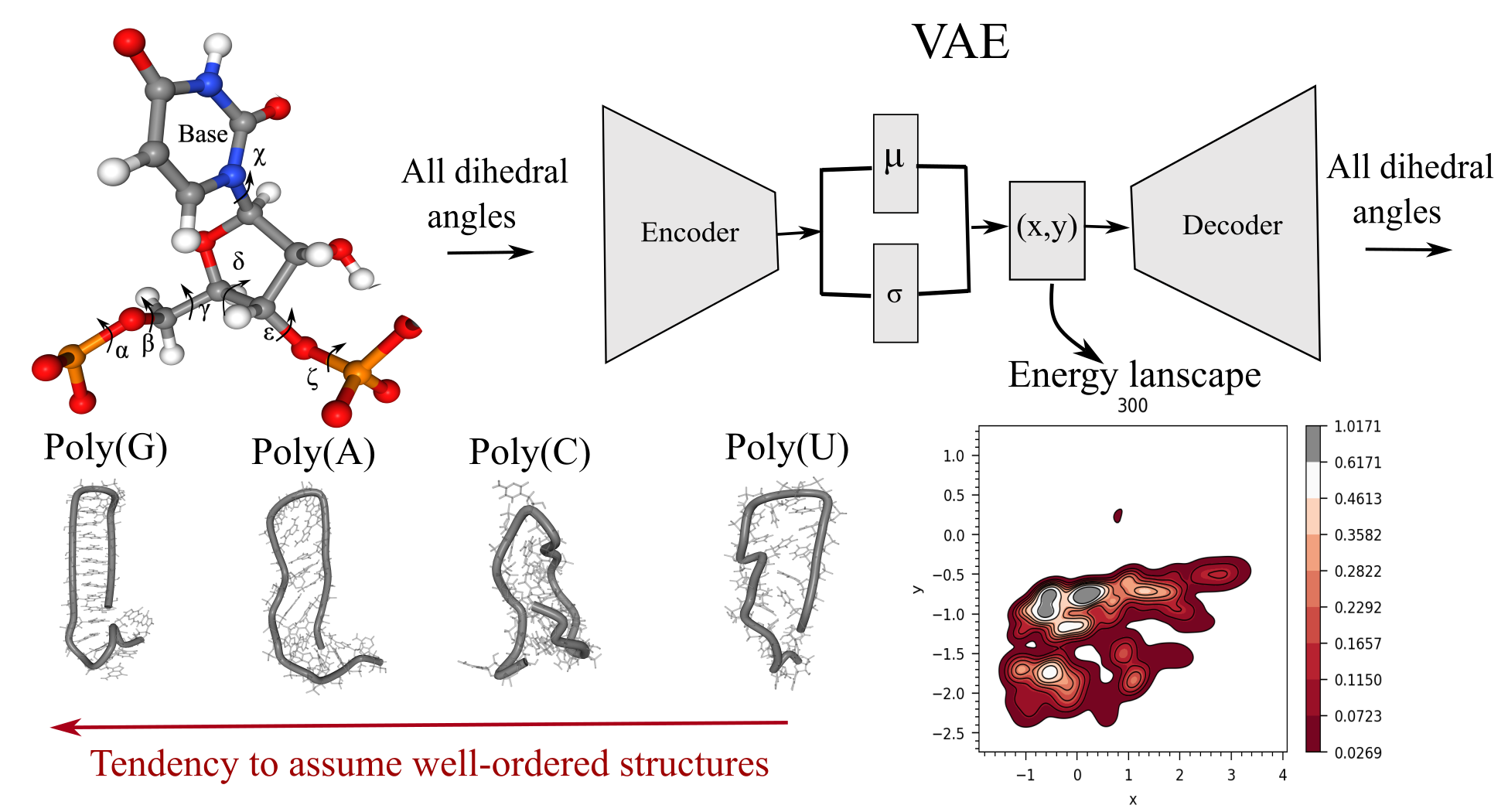
[43] Mapping energy landscapes of homopolymeric RNAs via simulated tempering and deep unsupervised learning
V Ramachandran, DA Potoyan*
Biophys J (2024)
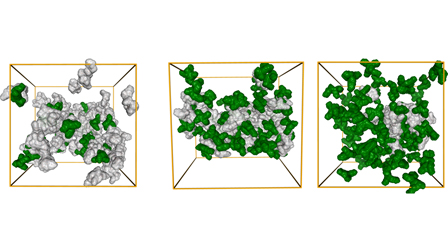
[42] Atomistic insights into the reentrant phase-transitions in
polyuracil and polylysine mixtures
V Ramachandran, DA Potoyan*
J Chem Phys (2024)
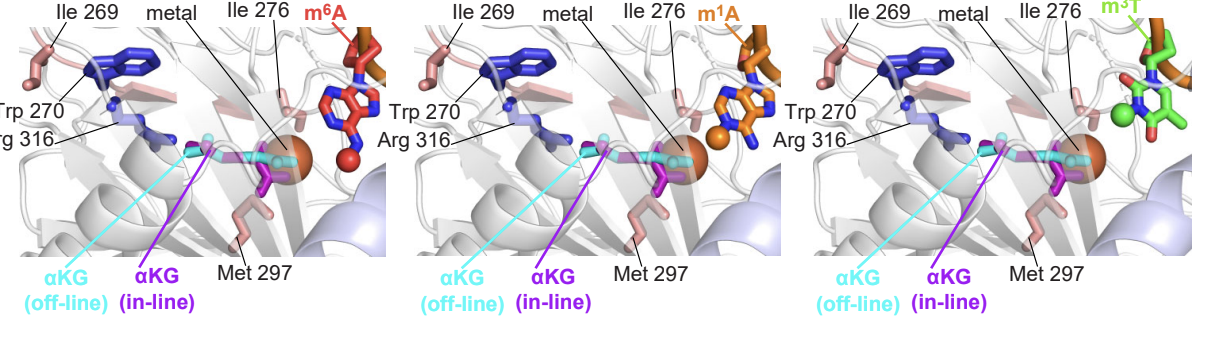
[41] An α-ketoglutarate conformational switch controls iron accessibility,
activation, and substrate selection of the human FTO protein
D Burns, B Khatiwada, A Singh, JA Purslow, DA Potoyan* V Venditti*
Proc Natl Acad Sci (2024)
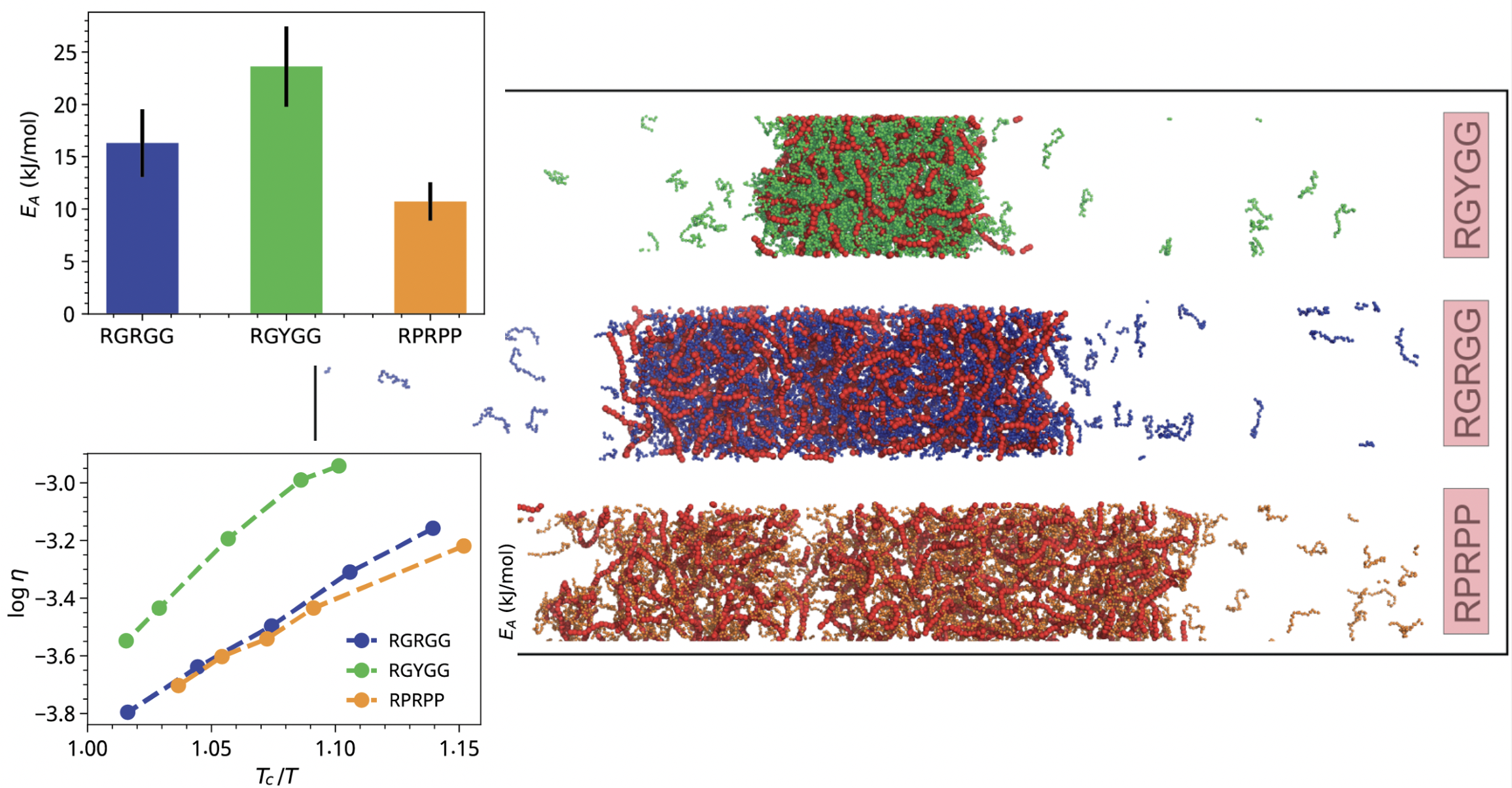
[40] Determinants of Viscoelasticity and Flow Activation Energy in Biomolecular Condensates
I Alshareedah, A Singh, S Yang, V Ramachandran, A Quinn,
DA Potoyan*, PR Banerjee*
Sci Adv (2024)
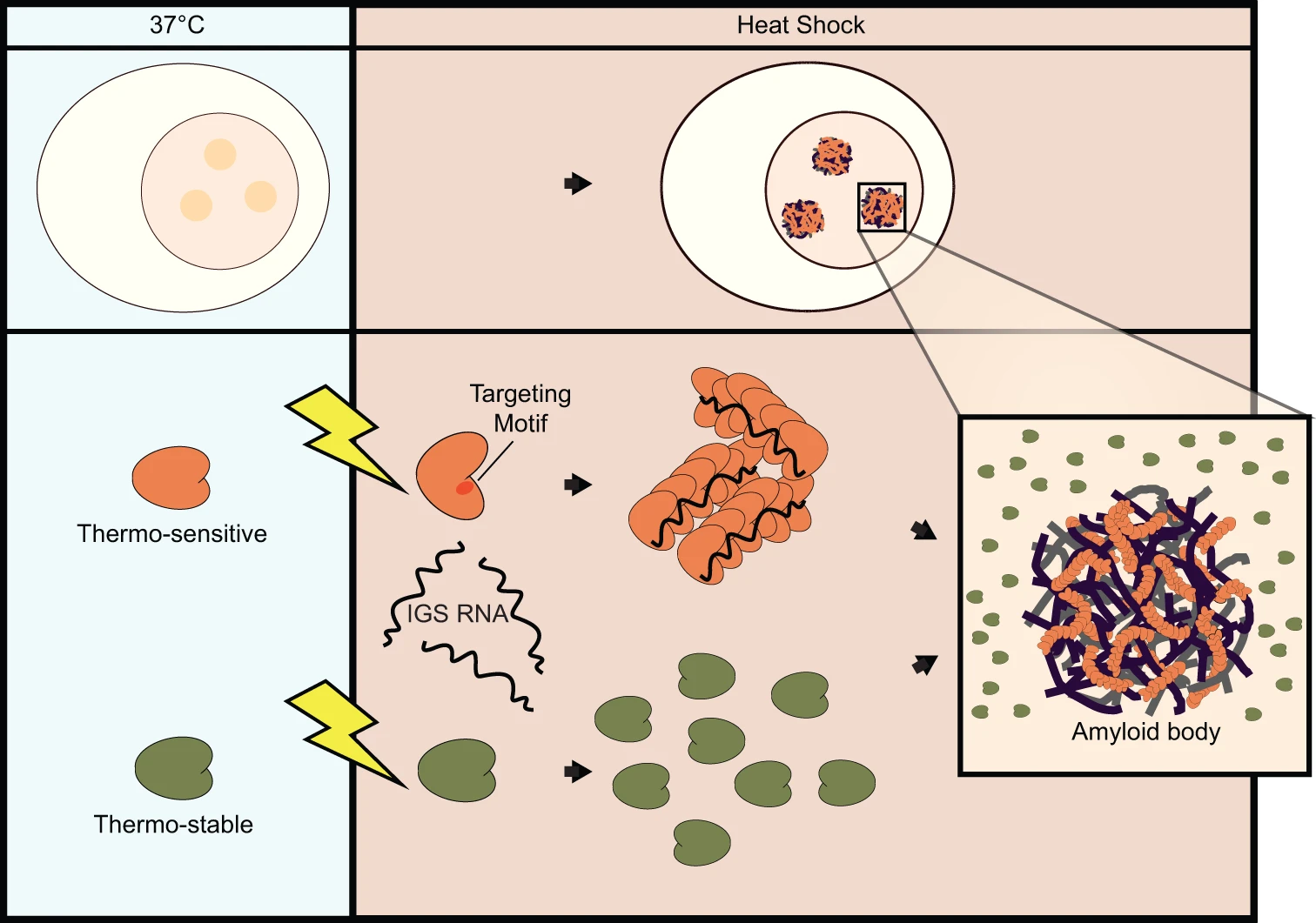
[39] Protein thermal sensing regulates physiological amyloid aggregation
D Marijan, EA Momchilova, D Burns, S Chandhok, R Zapf, H Wille,
DA Potoyan, TE Audas
Nat Comm (2024)
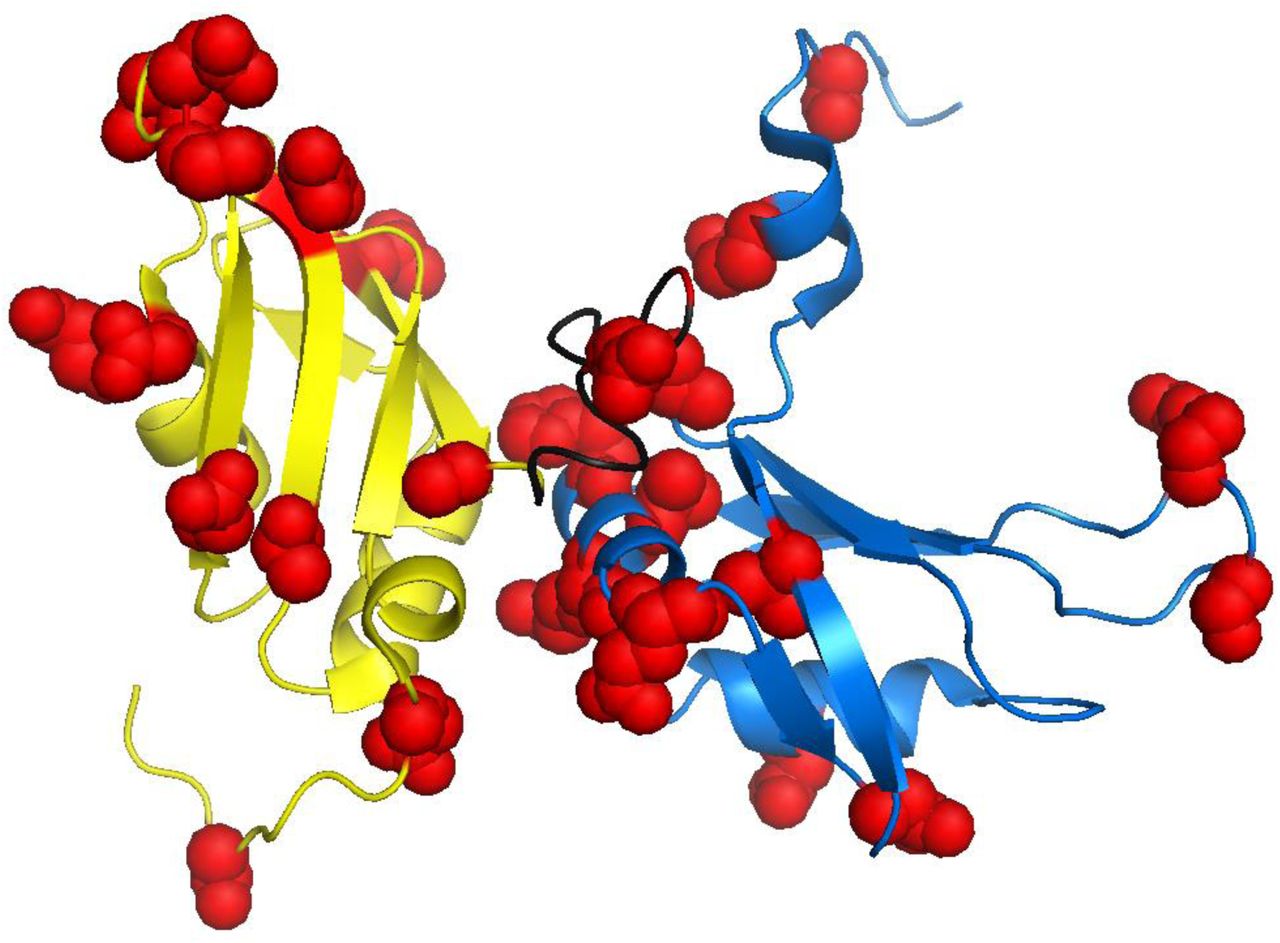
[38] Thermodynamic Coupling of the tandem RRM domains of hnRNP A1 underlie its Pleiotropic RNA Binding Functions
JD Levengood, DA Potoyan, S Penumutchu, A Kumar, Y Wang, AL Hansen, S Kutluay, J Roche, Blanton S Tolbert
Sci Adv (2024)
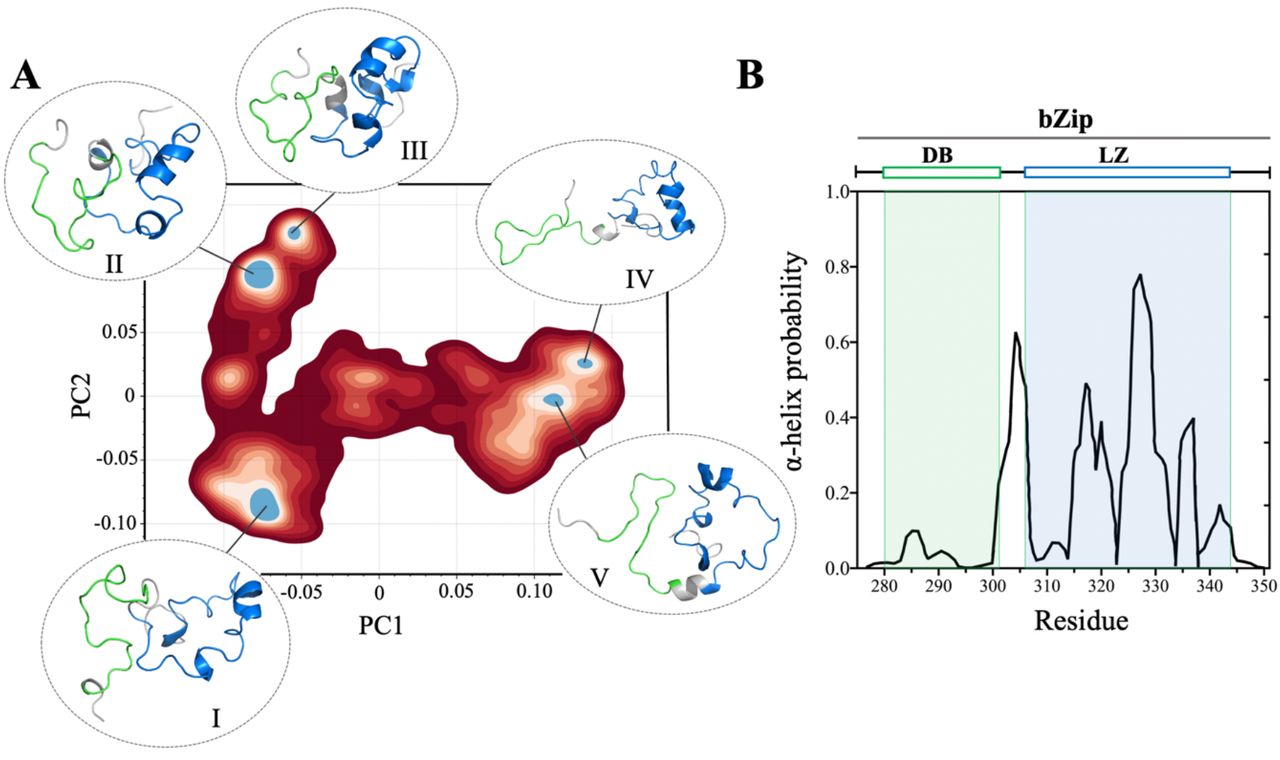
[37] Conformational landscape of the transcription factor ATF4 is dominated by disordered-mediated inter-domain coupling
U Patel, S Siang, DA Potoyan, J Roche
Biochemistry (2024)
[36] An interdisciplinary effort to integrate coding into science courses. Nature Computational Science
Vizcarra, C. L. et al; Nat Comp Sci (2024)
(2023)
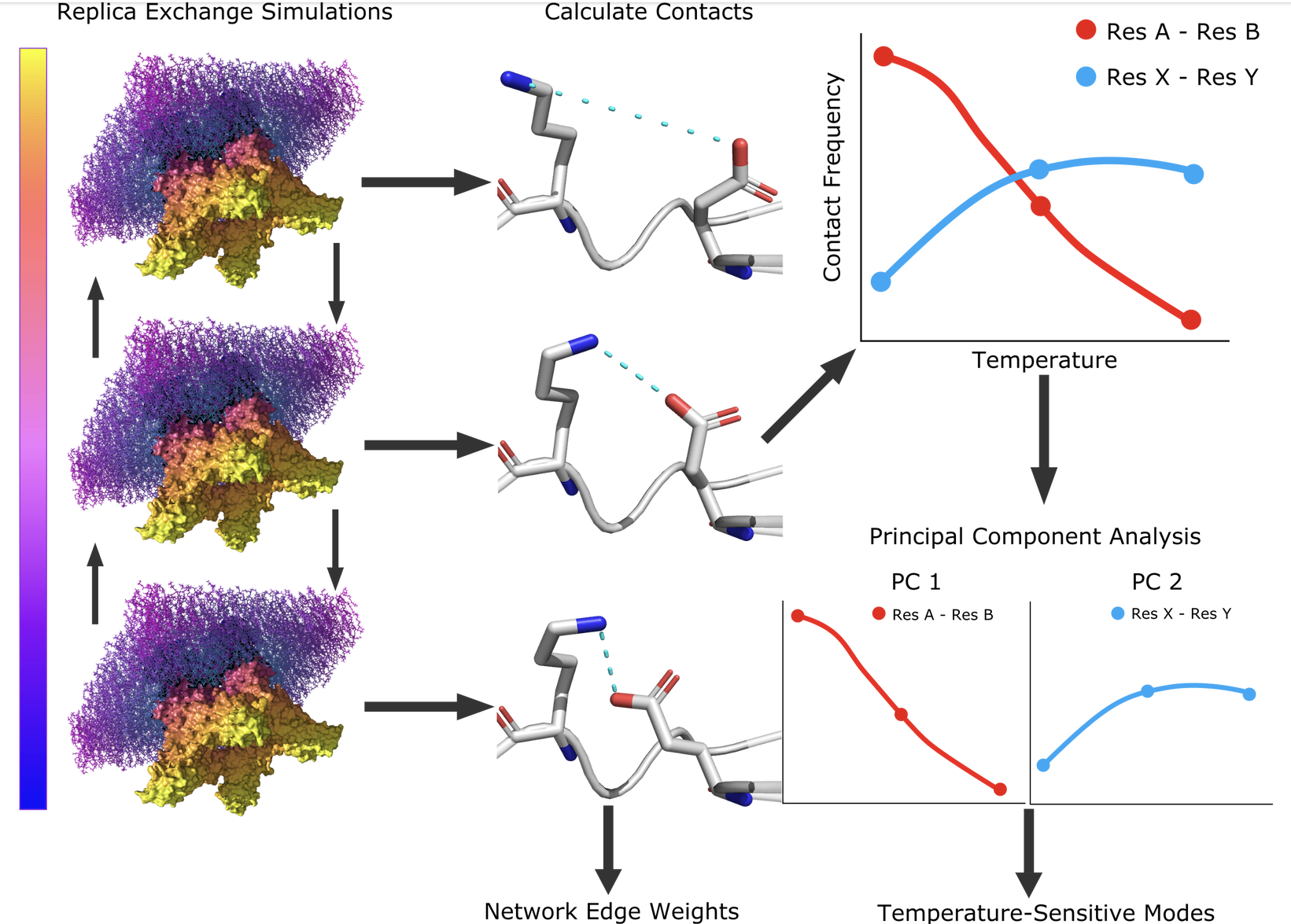
[35] Temperature-Sensitive Contact Modes Allosterically Gate TRPV3
D Burns, V Venditti*, DA Potoyan*
Plos Comp Bio (2023)
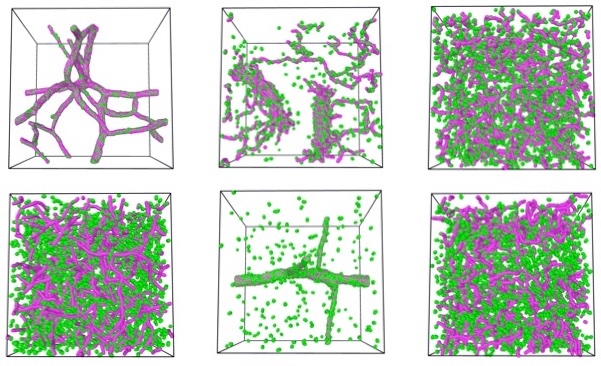
[34] Re-entrant transitions of locally stiff RNA chains in the presence of polycations
leads to gelated architectures
I Malhotra, DA Potoyan*
Soft Matter (2023)
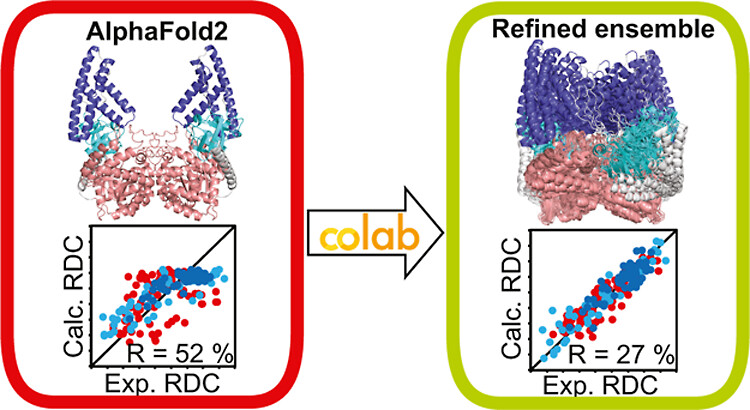
[33] Solution Structure Ensembles of the Open and Closed Forms
of the ∼130 kDa Enzyme I via AlphaFold Modeling, Coarse Grained Simulations, and NMR
A Singh, S Sedinkin, D Burns, D Shukla, DA Potoyan*, V Venditti*
J Am Chem Soc (2023)
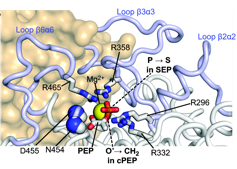
[32] Protein conformational dynamics underlay selective recognition of
thermophilic over mesophilic Enzyme I by a substrate analogue
A Singh, D Burns, S Sedinkin, B Van Veller, DA Potoyan*, V Venditti*
Biomolecules (2023)
(2022)
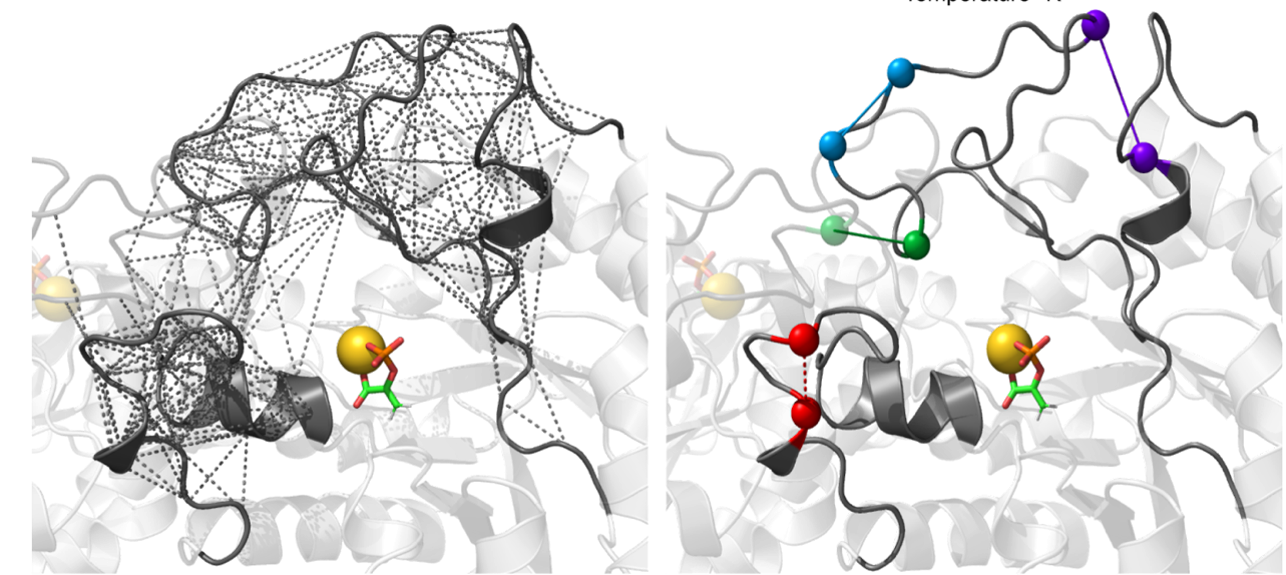
[31] Temperature Sensitive Contacts in Disordered Loops Tune Enzyme I Activity
D Burns, A Singh, V Venditti*, DA Potoyan*;
Proc Natl Acad Sci (2022)
[30] On the role of stoiciometry, salt and external crowding in protein-RNA
phase separation
R Laghmach, I Alshareedah, M Pham, M Raju, P Banerjee*, DA Potoyan*;
iScience (2022)

[29] BIAPSS: A comprehensive physicochemical analyzer of proteins
undergoing liquid-liquid phase separation
AE Badaczewska-Dawid, VA Uversky*; DA Potoyan*;
Int J Mol Sci (2022)
[28] A Liquid State Perspective on Dynamics of Chromatin Compartments
R Laghmach, M Di Pierro DA Potoyan*;
Front. Mol. Biosci. (2022)
[27] Multi-scale models of protein-RNA phase separation
R Laghmach, I Malhotra, DA Potoyan*;
Methods in Molecular Biology (2022)
(2021)
[26] The interplay of chromatin phase separation and lamina interactions in nuclear organisation
R Laghmach, M Di Pierro DA Potoyan*;
Biophys J (2021)
[25] Programmable Viscoelasticity in Protein-RNA Condensates with Disordered
Sticker-Spacer Polypeptides
I Alshareedah, M M Moosa, M Pham, DA Potoyan*, PR Banerjee* Nat Comm (2021)
[24] Solvent Exposure and Ionic Condensation Drive Fuzzy Dimerization of Disordered Heterochromatin Protein Sequence
J Mueterthies, DA Potoyan*;
Biomolecules (2021)
[23] Sequence-encoded and Composition-dependent Protein-RNA Interactions Control Multiphasic Condensate Topologies
T Kaur, M Raju, I Alshareedah, RB Davis, DA Potoyan*, P Banerjee*;
Nat Comm (2021)
(2020)
[22] Liquid-liquid phase separation driven compartmentalization of reactive nucleoplasm
R Laghmach, DA Potoyan*;
Phys Biol (2020)
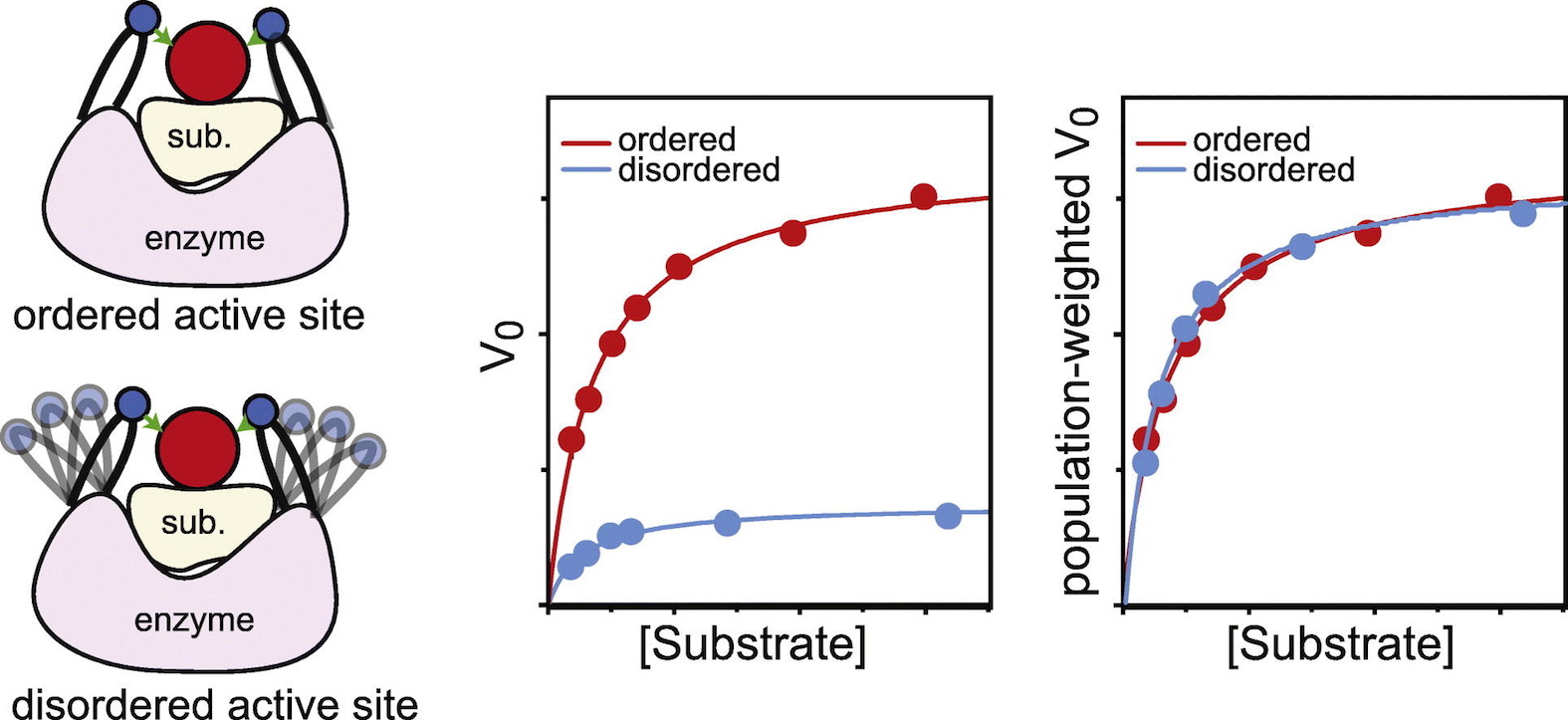
[21] Hybrid thermophilic/mesophilic enzymes reveal a role
for conformational disorder in regulation of bacterial Enzyme I
RR Dotas, TT Nguyen, CE Stewart Jr, R Ghirlando,
DA Potoyan*, V Venditti*;
J Mol Biol (2020)
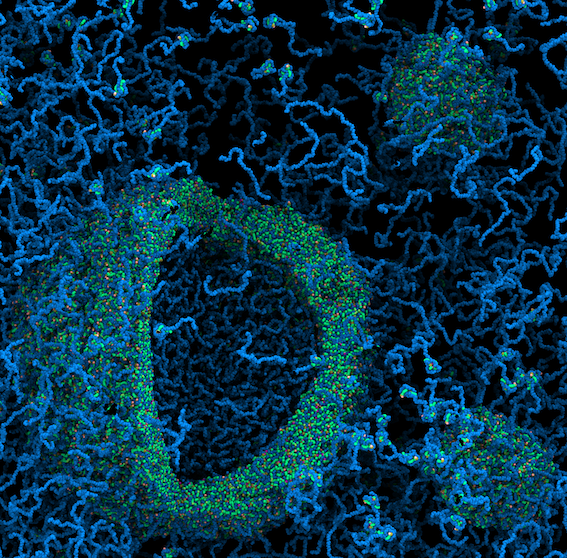
[20] Phase Transition of RNA-protein Complexes into Ordered Hollow Condensates
I Alshareedah, MM Moosa, M Raju, DA Potoyan*, P Banerjee*;
Proc Natl Acad Sci (2020)
(2019)
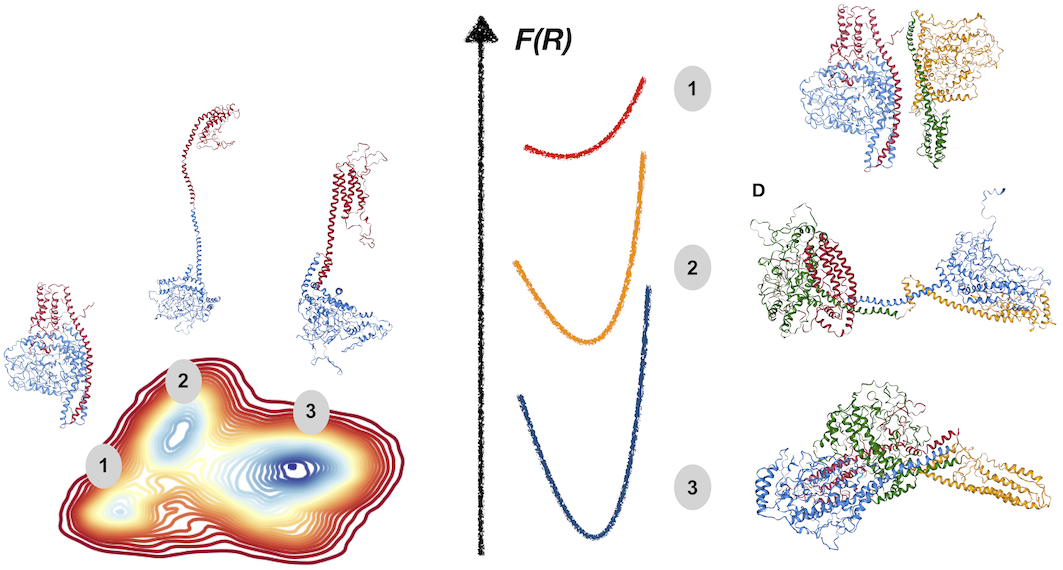
[19] Disorder mediated oligomerization of DISC1 proteins revealed by coarse-grained computer simulations.
J Roche, DA Potoyan
J Phys Chem B (2019)
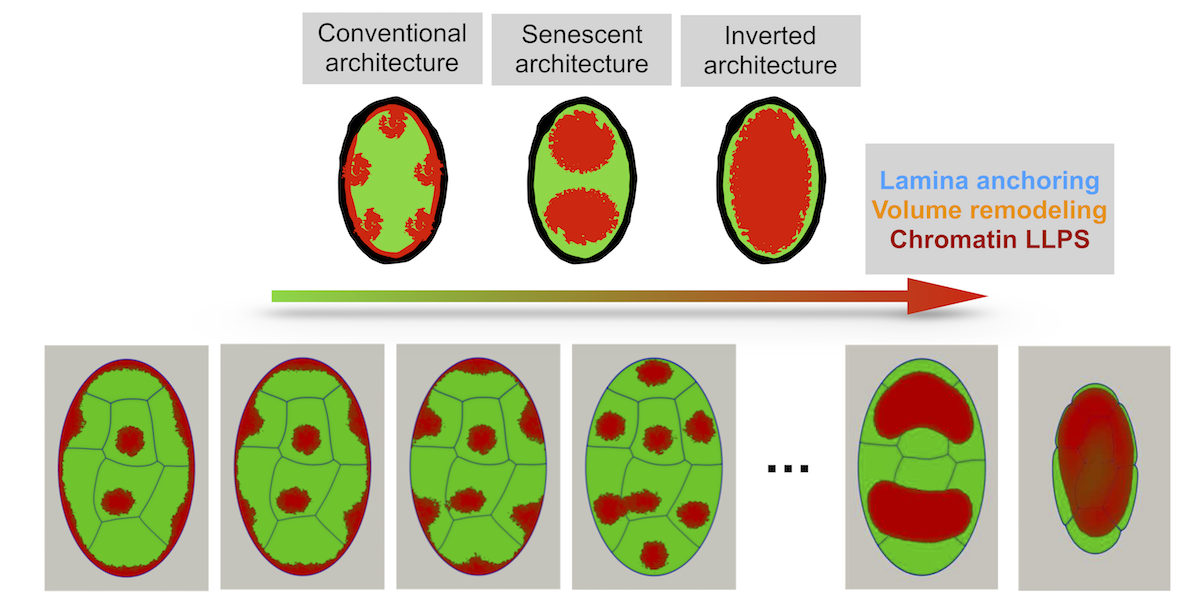
[18] Mesoscale Liquid Model of Chromatin Recapitulates Nuclear Order of Eukaryotes.
R Laghmach, M Di Pierro, DA Potoyan
Biophys J (2019)
(2018)
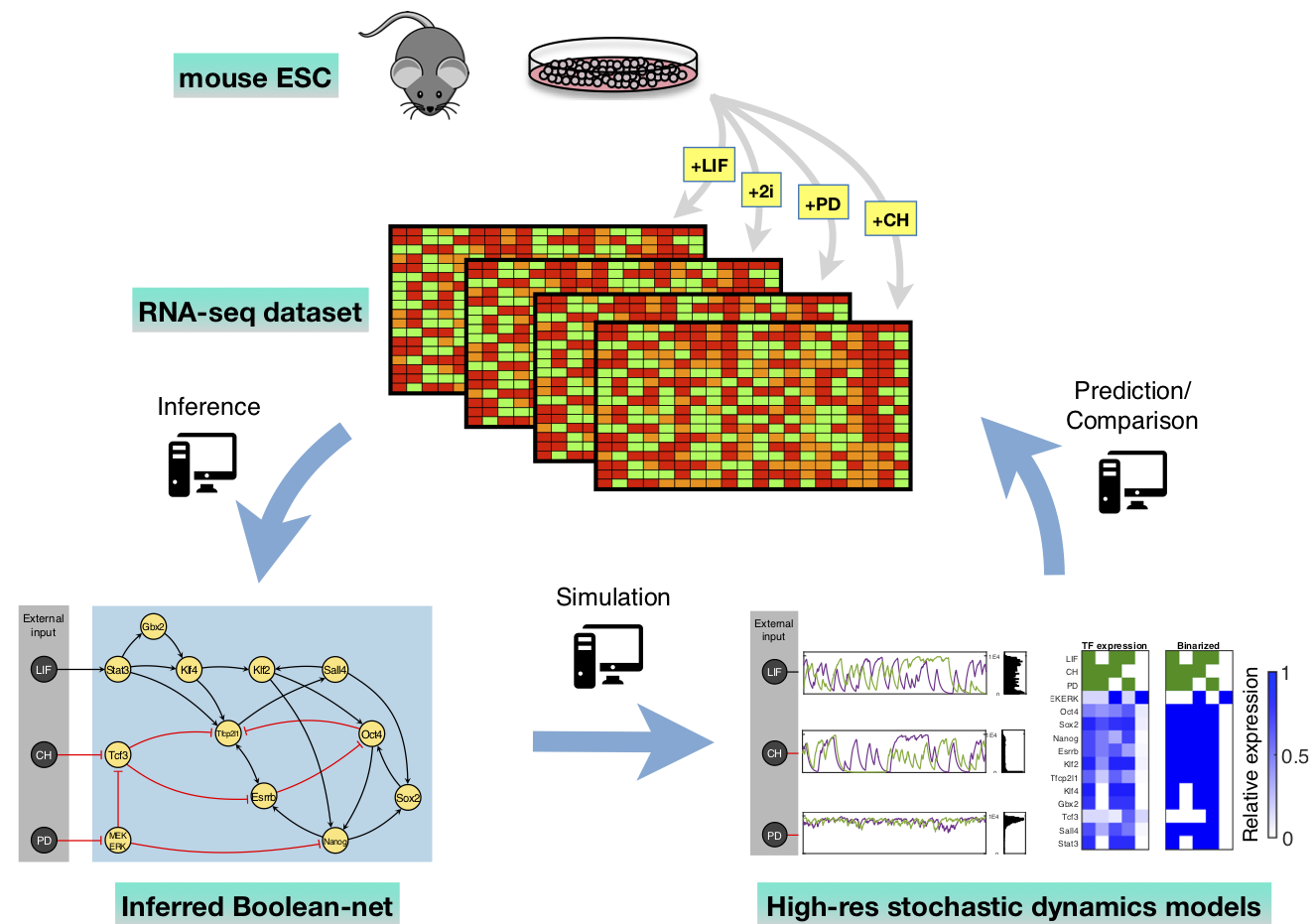
[17] A stochastic and dynamical view of pluripotency in mouse embryonic stem cells.
YT Lin, PG Hufton, EJ Lee, DA Potoyan
PLoS. Comp. Bio. (2018)
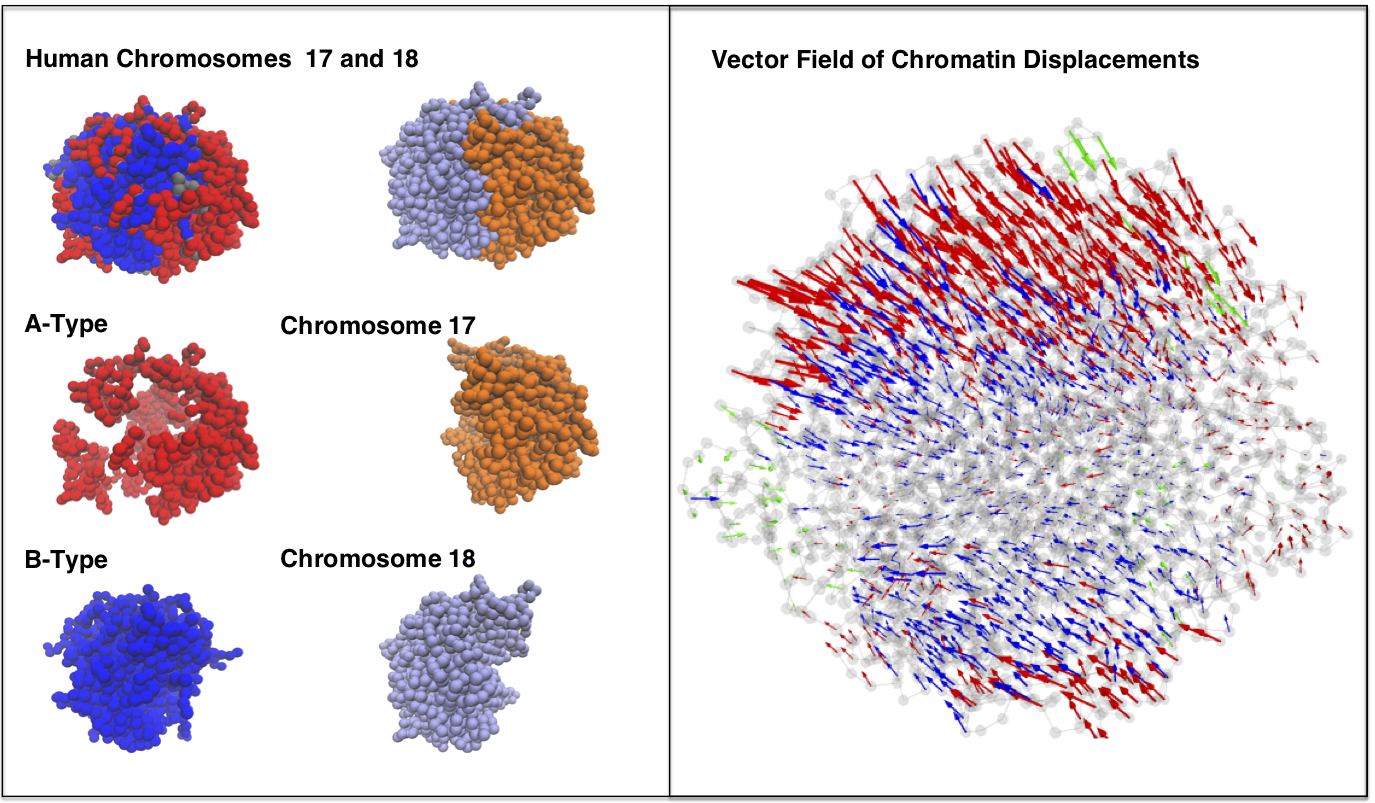
[16] Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes.
M Di Pierro*, DA Potoyan* , PG Wolynes, JN Onuchic
Proc. Natl. Acad. Sci. (2018)
Work done prior to ISU (2010-2017)
[15] Modeling the therapeutic efficacy of NFkB synthetic decoy oligodeoxynucleotides.
Z Wang, DA Potoyan, PG Wolynes; BMC Sys. Biol. (2018)
[14] Stochastic Resonances in a Distributed Genetic Broadcasting System: The NFκB/IκB paradigm.
Z Wang*, DA Potoyan*, PG Wolynes; J. Roy. Soc. Int. (2018)
[13] Resolving the NFkB Heterodimer Binding Paradox: Strain and Frustration Guide the Binding of Dimeric Transcription Factors
DA Potoyan, C Bueno, W Zheng, EA Komives, PG Wolynes; J. Am. Chem. Soc. (2017)
[12] Stochastic dynamics of genetic broadcasting networks
DA Potoyan, PG Wolynes; Phys. Rev. E. (2017)
[11] Molecular stripping, targets and decoys as modulators of oscillations in
the NFκB/IκBα/DNA genetic network
Z Wang*, DA Potoyan*, PG Wolynes; J. Roy. Soc. Int. (2016)
[10] PEST control of molecular stripping of NFkB from DNA transcirption sites
DA Potoyan, W Zheng, DU Fereiro, PG Wolynes, EA Komives; J. Phys. Chem. B (2016)
[9] Molecular stripping in the NFkB-IkB-DNA genetic switch
DA Potoyan, W Zheng, EA Komives, PG Wolynes; Proc. Natl. Acad. Sci. (2016)
[8] Dichotomous noise based models of genetic switching
DA Potoyan, PG Wolynes; J. Chem. Phys. (2015)
[7] The acetylation landscape of the H4 histone tail: disentangling the interplay between the specific and cumulative effects.
D Winogradoff, I Echeverria, DA Potoyan, GA Papoian; J. Am. Chem. Soc. (2015)
[6] On the dephasing of genetic oscillators
DA Potoyan, PG Wolynes; Proc. Natl. Acad. Sci. (2014)
[5] Recent successes in coarse-grained modeling of DNA
DA Potoyan, A Savelyev, G Papoian; WIRES Comp. Mol. Sci. (2013)
[4] Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation
DA Potoyan, G Papoian; Proc. Natl. Acad. Sci. (2012)
[3] Computing Free Energy of a Large-Scale Allosteric Transition in Adenylate Kinase Using All Atom Explicit Solvent Simulations
DA Potoyan, P Zhuravlev, G Papoian; J. Phys. Chem. B (2012)
[2] Energy Landscape Analyses of Disordered Histone Tails Reveal Special
Organization of Their Conformational Dynamics.
DA Potoyan, G Papoian; J. Am. Chem. Soc. (2012)
[1] Computing free energies of protein conformations from explicit solvent simulations.
P Zhuravlev, S Wu, DA Potoyan, M Rubinstein, G Papoian; Methods (2010)